The purpose of this database is to represent the relationship between protein structural change and ligand binding. We classified protein structural changes into 7 classes blow, in terms of the ligand binding sites and the location where the dominant motion occurs.
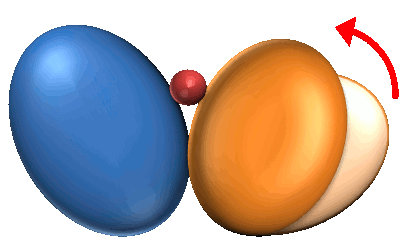 |
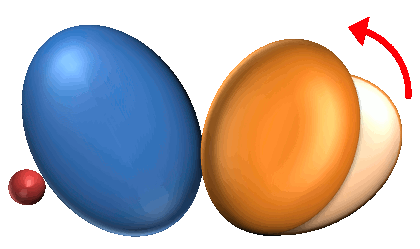 |
1. Coupled Domain Motion (CD) : 59 |
2. Independent Domain Motion (ID) : 70 |
 |
 |
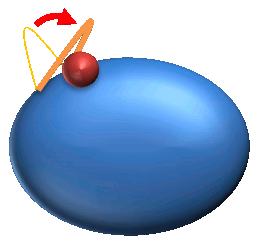
3. Coupled Local Motion (CL) : 125 |
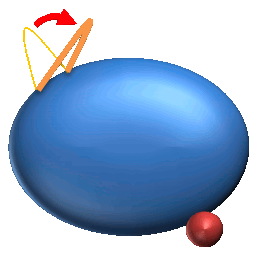
4. Independent Local Motion (IL) : 135 |
 |
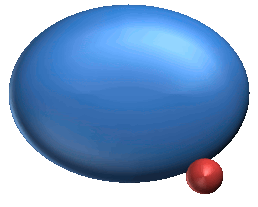 |
5. Burying Ligand Motion (B) : 104 |
6. No Significant Motion (N) : 311 |
 |
|
7. Other Motion (O) : 35 |
|
1. Coupled Domain motions are the domain motions induced upon ligand binding.
2. Independent Domain motions are the observable domain motions regardless of ligand binding.
3. Coupled Local motions are the local motions induced upon ligand binding.
4. Independent Local motions are the observable local motions regardless of ligand binding.
5. Burying ligand motions are imaginable motions required to hold ligand protein-inside.
6. No significant motions mean just nothing happen.
7. Other motions are motions unclassified into domain and local motions.
References
|
1. |
T. Amemiya, R. Koike, S. Fuchigami, M. Ikeguchi, and A. Kidera. |
|
|
|
This database was created by T. Amemiya. Please contact by e-mail at
The Background information in PSCDB are available.
